PDC 2022 is the 2022 edition of the Process Discovery Contest.
There can be only one…
Do you believe you have implemented a good discovery algorithm?
Then submit it to the PDC 2022 to put it to the test!
 The PDC 2022 test contains 480 event logs, for which a model needs to be discovered (training logs), and 96 pairs of event logs that are used to evaluate the discovered models (test logs and base logs) .
The PDC 2022 test contains 480 event logs, for which a model needs to be discovered (training logs), and 96 pairs of event logs that are used to evaluate the discovered models (test logs and base logs) .
The 96 pairs of test logs and base logs are all generated using the same configurable model, which was inspired by a model from a real-life event log and has the following configurable options:
- Long-term dependencies: yes/no
- Loops: no/simple/complex
- A simple loop has a single point of entry and a single point of exit.
- A complex loop has multiple points of entry and/or multiple points of exit.
- OR constructs: yes/no
- Routing constructs: yes/no
- Optional tasks: yes/no
- Duplicate tasks: yes/no
Each pair is matched by five training logs:
- A training log without noise.
- A training log where in every trace with probability 20% one random event is removed.
- A training log where in every trace with probability 20% one random event is moved to a random position.
- A training log where in every trace with probability 20% one random event is copied to a random position.
- A training log where in every trace with probability 20% either one random event is removed (40%), moved (20%), or copied (40%).
Each training log contains 1000 traces that result from random walks through the configured model. Each test log and each base log contains 1000 traces. For every pair of a test log and a base log we determine for every trace from the test log whether it fits the discovered model better than the corresponding trace from the base log. 500 traces from the test log fit the original model better than the corresponding trace from the base log, and 500 do not. For sake of completeness: Two traces from both logs correspond if and only if they are at the same position in the corresponding logs: The fifth trace of the test log is classified against the fifth trace of the base log, etc.
How, what and when to submit?
Please let Eric Verbeek know that you want to submit your implemented discovery algorithm. Eric will then provide you with a link where you can upload your submission.
You should submit a working discovery algorithm, which can be called using a “Discover.bat” Windows batch file which takes two parameters:
- The full path to the training log file, excluding the “.xes” extension.
- The full path to the model file where the discovered model should be stored, excluding any extension like “.pnml”, “.bpmn”, or “.lsk”.
As an example, assuming that the miner discovers a Petri net,
Discover.bat logs\discovery\discovery-log models\discovered-model
will discover a model from the training log file “logs\discovery\discovery-log.xes” and will export the discovered Petri net to the file “models\discovered-model.pnml”.
If the results of calling your Discovery.bat file as described above is a PNML file (Petri nets), a BPMN file (BPMN diagram), or a LSK file (log skeleton), then you’re done. If not, the discovery algorithm needs to come with its own working classifier algorithm, that is, a “Classify.bat” Windows batch file, which takes four parameters:
- The full path to the test log file, excluding the “.xes” extension.
- The full path to the base log file, excluding the “.xes” extension.
- The full path to the model file which should be used to classify the test log, excluding any extension like “.pnml”, “.bpmn”, or “.lsk”.
- The full path to the log file where the classified test log should be stored, excluding the “.xes” extension.
As an example, assuming that the miner discovered a Petri net,
Classify.bat logs\test\test-log logs\base\base-log models\discovered-model logs\classified\test-log
will classify whether every trace from the test log “logs\test\test-log.xes” fits the Petri net from “models\discovered-model.pnml” better than the corresponding trace from the base log “logs\base\base-log.xes” and it will export the classified traces (by adding the “pdc:isPos” attributes to every trace) as an event log in “logs\classified\test-log.xes”.
Classification of a trace is done by adding the boolean “pdc:isPos” attribute to the trace, which should be true if the trace is classified positive (fits your model better than the corresponding trace in the base model) and false if the trace is classified negative (does not fit your model better than the corresponding trace in the base model).
Score and winners
For each training log, the “Discovery.bat” file is used to discover a model from the training log. Next, the “Classify.bat” is used to classify every trace in the test log against the corresponding trace in the base log using the discovered model. This results in a positive accuracy rate P and a negative accuracy rate N for this training log. From these, its F-score F is computed as 2*(P*N)/(P+N). The end score for the discovery algorithm is the average F-score over all 480 training logs.
The winner is the submission with the best end score.
Key Dates
| Submission Deadline | September 30, 2022 |
| Disclosure of the Data Set | October 1, 2022 |
| Winner Notification | October 7, 2022 |
| Winner Announcement | During ICPM 20222 |
Example discovery algorithms
We have run the three base miners on the data set: The Flower miner, the Directly Follows miner, and the Trace (or Sequence) miner.
Flower miner
The Flower miner scores 0%, with a positive accuracy of 0.0% and a negative accuracy of 100.0%.
The Flower miner scores 0% as every trace fits the discovered model. As a result no trace fits better than another trace.
Directly Follows miner
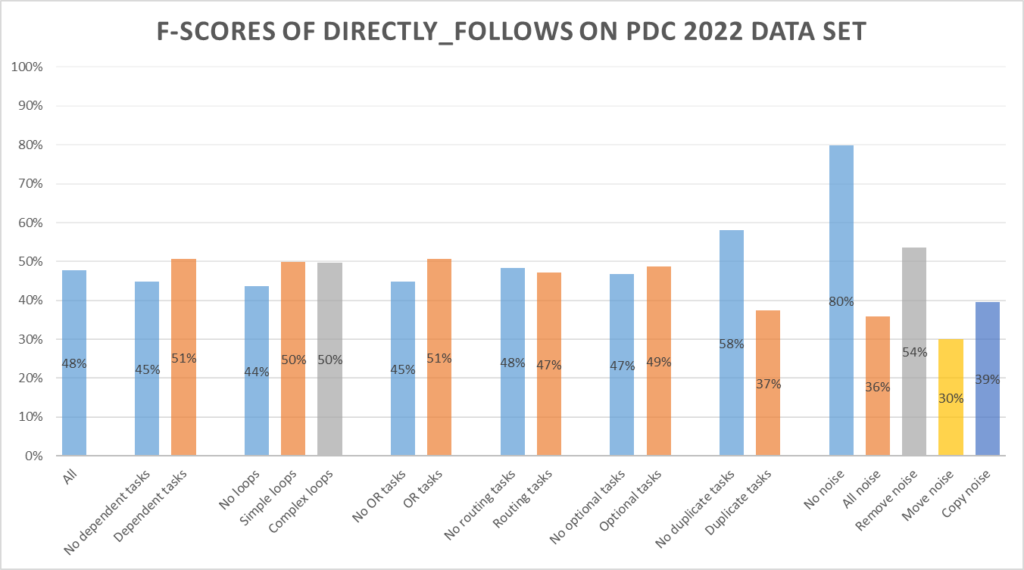
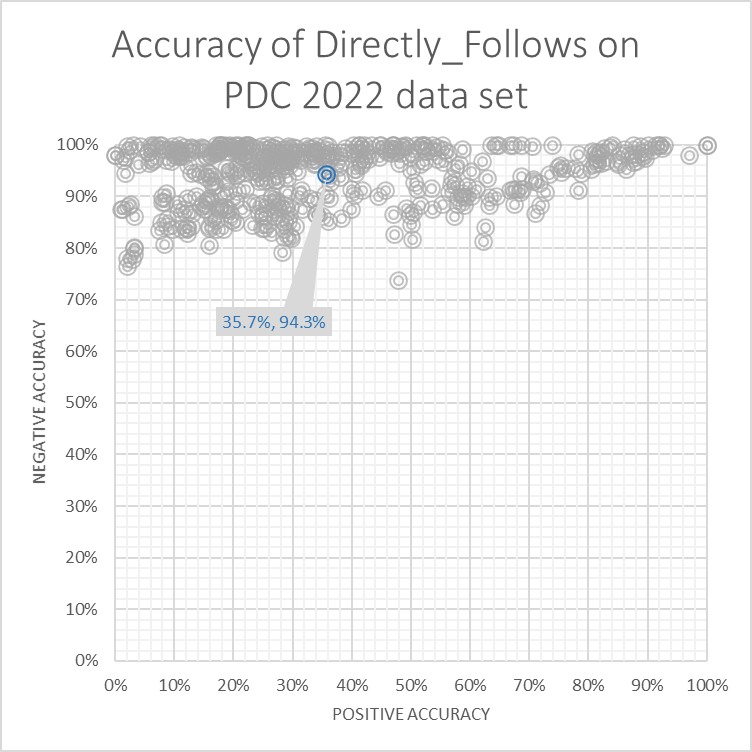
The Directly Follows miner scores 48%, with a positive accuracy of 35.7% and a negative accuracy of 94.3%.
Trace miner
The Trace miner scores 90%, with a positive accuracy of 86.0% and a negative accuracy of 95.3%.
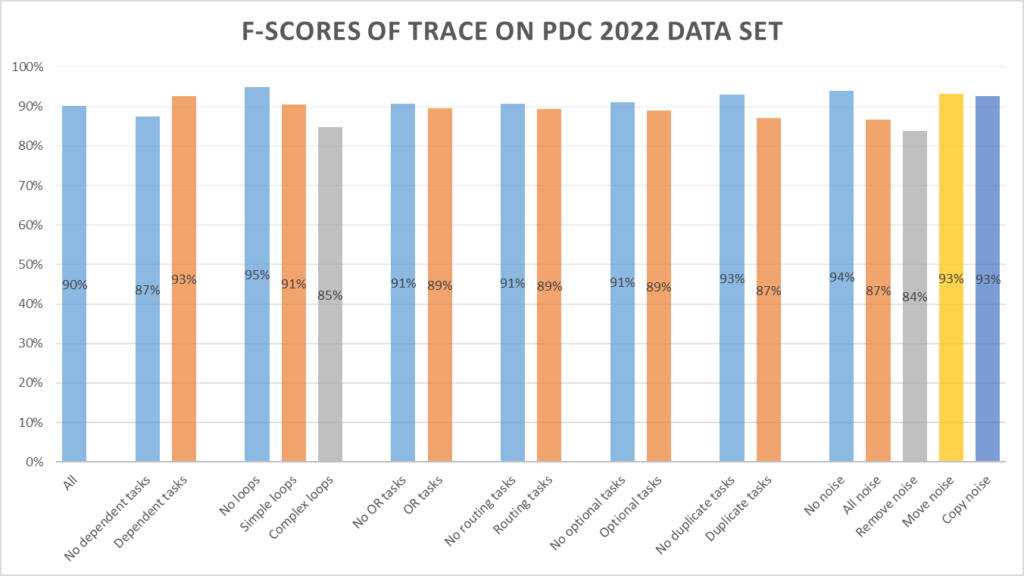
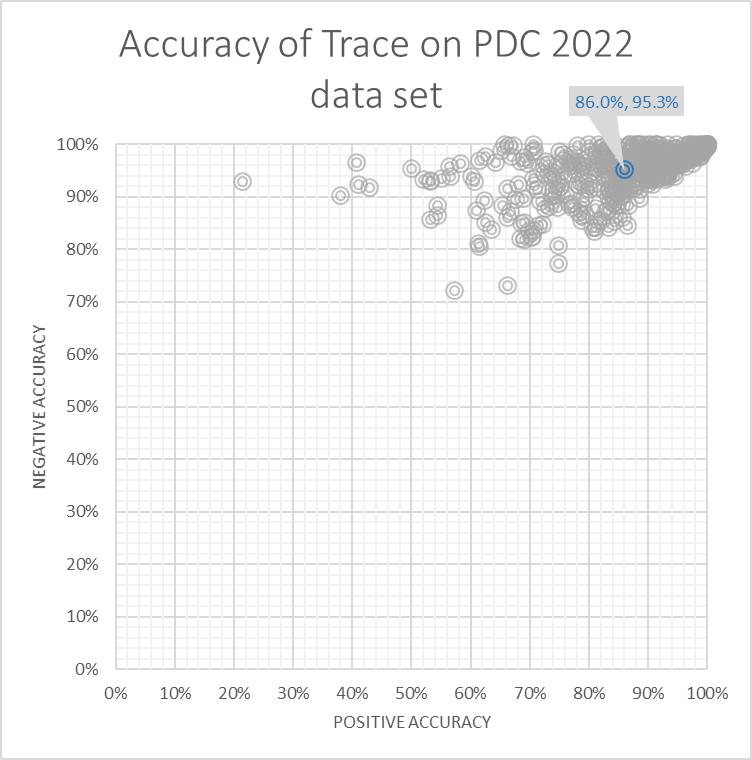
Submissions
The following twenty-seven algorithms were submitted:
- DisCoveRJS, by Tijs Slaats and Axel Christfort
- DisCoveRJS-NoiseFiltering, default configuration (0.2), by Tijs Slaats and Axel Christfort
- DisCoveRJS-NoiseFiltering, configuration 0.22, by Tijs Slaats and Axel Christfort
- DisCoveRJS-NoiseFiltering, configuration 0.23, by Tijs Slaats and Axel Christfort
- DisCoveRJS-NoiseFiltering, configuration 0.24, by Tijs Slaats and Axel Christfort
- DisCoveRJS-NoiseDetection, default configuration (0.990), by Tijs Slaats and Axel Christfort
- DisCoveRJS-NoiseDetection, configuration 0.985, by Tijs Slaats and Axel Christfort
- DisCoveRJS-NoiseDetection, configuration 0.980, by Tijs Slaats and Axel Christfort
- DisCoveRJS-NoiseDetection, configuration 0.975, by Tijs Slaats and Axel Christfort
- DisCoveRJS-ProbabilisticClassification, default configuration , by Tijs Slaats and Axel Christfort
- DisCoveRJS-ProbabilisticClassification, configuration invert, by Tijs Slaats and Axel Christfort
- NoiseFiltering, by Tijs Slaats, Paul Cosma and Axel Christfort
- UiPath_PIM_PDC2022, configuration A, by Dennis Brons
- UiPath_PIM_PDC2022, configuration B, by Dennis Brons
- UiPath_PIM_PDC2022, configuration C, by Dennis Brons
- UiPath_PIM_PDC2022, configuration D, by Dennis Brons
- UiPath_PIM_PDC2022, configuration E, by Dennis Brons
- UiPath_PIM_PDC2022, configuration F, by Dennis Brons
- UiPath_PIM_PDC2022, configuration G, by Dennis Brons
- UiPath_PIM_PDC2022, configuration H, by Dennis Brons
- UiPath_PIM_PDC2022, configuration I, by Dennis Brons
- UiPath_PIM_PDC2022, configuration J, by Dennis Brons
- EnrichedCC, configuration v1, by Gemma Di Federico
- EnrichedCC, configuration v2, by Gemma Di Federico
- EnrichedCC, configuration v3, by Gemma Di Federico
- EnrichedCC, configuration v4, by Gemma Di Federico
- EnrichedCC, configuration v5, by Gemma Di Federico
Results
| Submission | Score | Positive Accuracy | Negative Accuracy |
|---|---|---|---|
| DisCoveRJS | 53.0% | 40.8% | 96.9% |
| DisCoveRJS-NoiseFiltering 0.20 | 68.3% | 57.1% | 93.1% |
| DisCoveRJS-NoiseFiltering 0.22 | 68.8% | 57.7% | 92.9% |
| DisCoveRJS-NoiseFiltering 0.23 | 68.9% | 57.8% | 92.8% |
| DisCoveRJS-NoiseFiltering 0.24 | 68.9% | 57.8% | 92.6% |
| DisCoveRJS-NoiseDetection 0.990 | 60.1% | 48.1% | 94.2% |
| DisCoveRJS-NoiseDetection 0.985 | 62.7% | 53.3% | 80.5% |
| DisCoveRJS-NoiseDetection 0.980 | 61.4% | 52.6% | 77.2% |
| DisCoveRJS-NoiseDetection 0.975 | 59.7% | 51.8% | 73.4% |
| DisCoveRJS-ProbablisticCLassification | 19.5% | 15.1% | 37.1% |
| DisCoveRJS-ProbablisticCLassification invert | 69.0% | 84.9% | 62.9% |
| NoiseFiltering | 85.0% | 83.2% | 90.1% |
| UiPath_PIM_PDC2022 A | 81.8% | 79.9% | 83.6% |
| UiPath_PIM_PDC2022 B | 81.8% | 80.4% | 83.3% |
| UiPath_PIM_PDC2022 C | 81.9% | 80.5% | 83.5% |
| UiPath_PIM_PDC2022 D | 84.4% | 82.8% | 86.1% |
| UiPath_PIM_PDC2022 E | 87.1% | 85.4% | 88.9% |
| UiPath_PIM_PDC2022 F | 89.2% | 87.3% | 91.2% |
| UiPath_PIM_PDC2022 G | 89.2% | 87.3% | 91.2% |
| UiPath_PIM_PDC2022 H | 89.3% | 87.4% | 91.5% |
| UiPath_PIM_PDC2022 I | 89.5% | 87.2% | 92.2% |
| UiPath_PIM_PDC2022 J | 89.5% | 87.2% | 92.2% |
| EnrichedCC v1 | 22.3% | 85.0% | 18.3% |
| EnrichedCC v2 | 31.8% | 69.9% | 29.5% |
| EnrichedCC v3 | 9.7% | 96.4% | 6.3% |
| EnrichedCC v4 | 28.4% | 77.0% | 25.5% |
| EnrichedCC v5 | 19.6% | 87.0% | 13.3% |
The positive accuracy is the percentage of positive traces correctly classified as positive, and the negative accuracy is the percentage of negative trace correctly classified as negative.
Winner
The best overall submission are the UiPath_PIM_PDC2022 configurations I and J, by Dennis Brons, which both score 89.5%. Congratulations to Dennis!
Downloads
Example discovery algorithms
- Directly follows miner
- ZIP archive, 295.3 MB
- Flower miner
- ZIP archive, 276.9 MB
- Trace miner
- ZIP archive, 511.5 MB
Classification algorithms
- BPMN Diagrams (BPMN)
- ZIP archive, 280.3 MB
- Log Skeletons (LSK)
- ZIP archive, 280.3 MB
- Petri nets (PNML)
- ZIP archive, 280.3 MB
Scorer algorithms
- Scorer
- ZIP archive, 280.3 MB
